Terokit manual for users
All of the data in TeroKit were collected from the public sources and mannully checked, however, there are still inevitable mistakes such as the wrong structures or wrong anannotations. User who find any mistakes are welcome to give us a feedback by email to qmclab@126.com and upload the right data to us by upload page.
Browe terokit
Users can browse terokit by compounds, scaffolds, bio-source, targets and vendors via the "Browse" button on the navigation bar on each page.
1. Browse by compounds
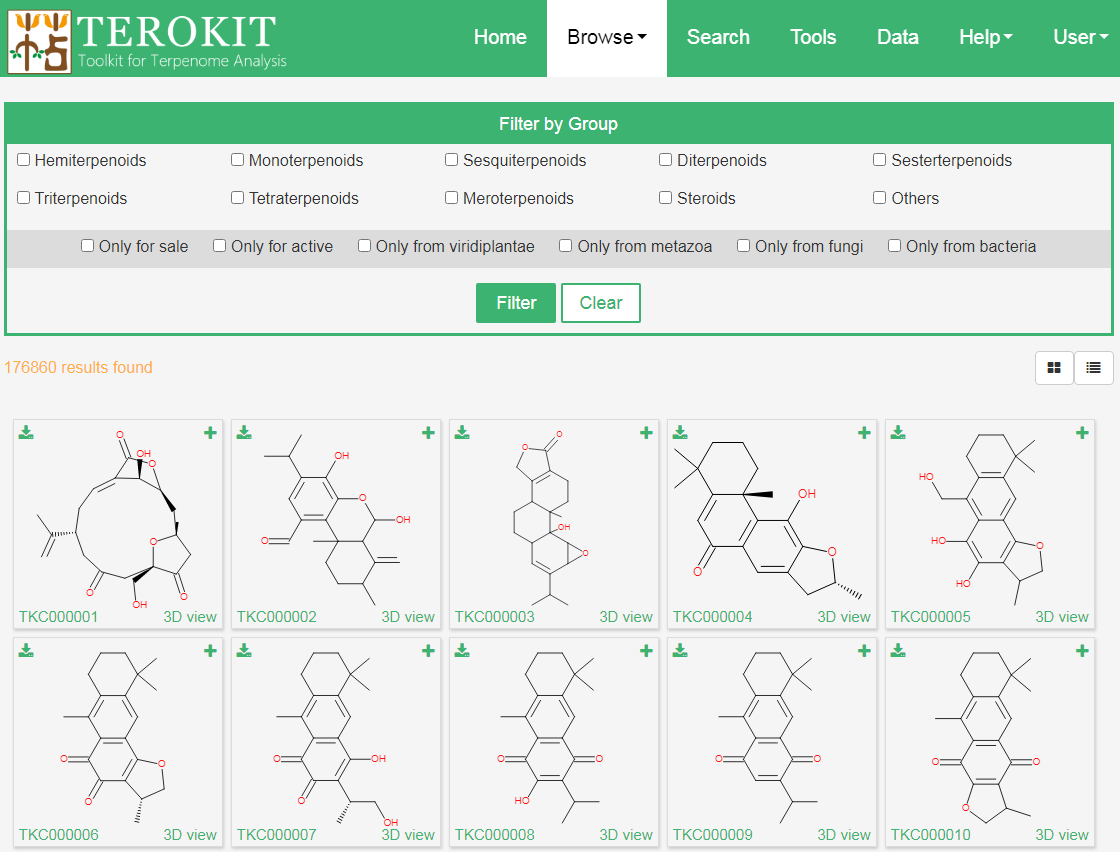
There are 30 molecules on the page at a time, users can click the buttons upper right the structure panel to switch to the information list. In the structure grid view (which is the default view) users can call out a modal window that shows some physicochemical and ADMET properties by clicking the “plus” button upper right each of the structure grid and upper right to download the sdf file of the compound. While click the compound ID that prefix with “TKC” will open a new page leading to the compound page showing the detailed information. The 3D structures are also provided.
Users can also browse compounds in different groups using the filter panel in the top.
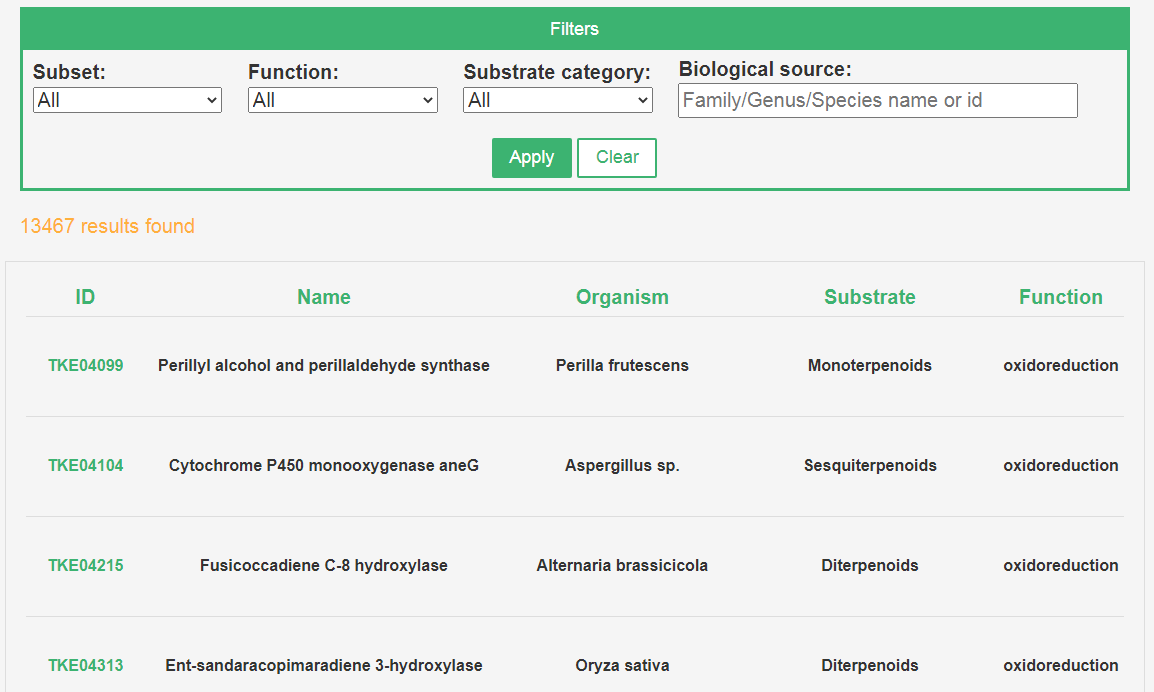
2. Browse by enzymes
Terpenome biosynthetic enzymes such as terpene synthases (TeroTPS) and cytochrome P450 monooxygenases and glycosyl transferase (TeroP450 and TeroGTS, under construction) can be browsed. The information such as protein name, organism, function and so on are list in the table, more details can be accessed by clicking the TPS ID in the first column. It should be noted that only terpene synthases (TeroTPS) is available only for the time being.
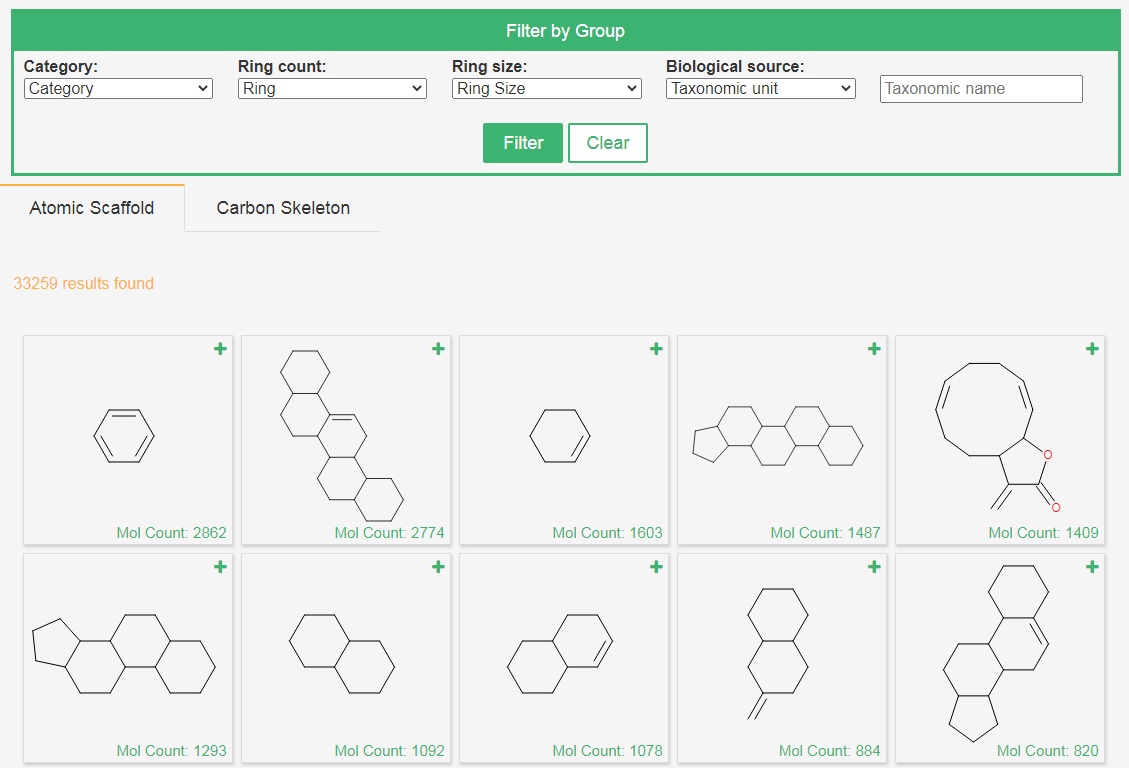
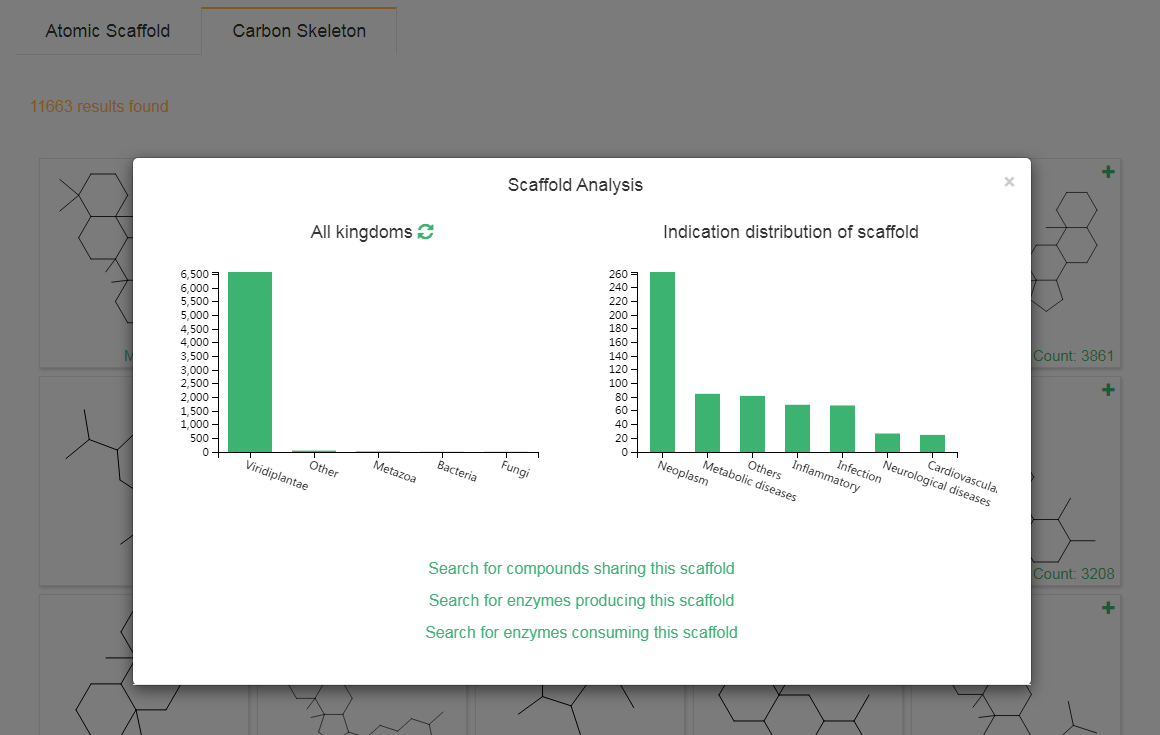
3. Browse by scaffolds
The Bemis-Murckoatomic scaffold and carbon skeleton of each compound are calculated by RDKit, the number of compounds sharing the same scaffold is showed and and users can get the distribution of organisms for each scaffold by clicking the “plus” button.
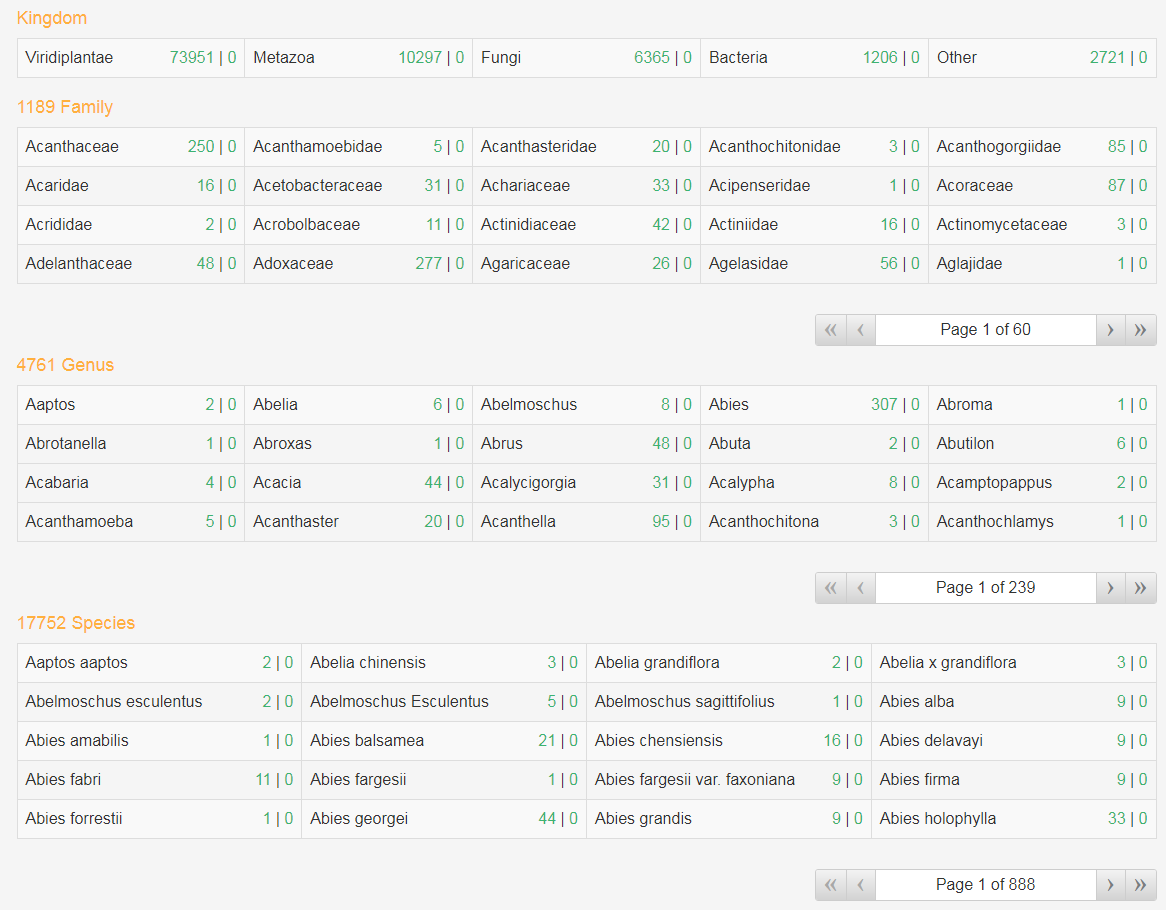
3. Browse by bio-source
The organism, family, genus and species of the source can be browsed, users can click the numbers behind the name of specific source to browse all compounds (the first one ) or enzymes (the second one) derived from it. And user can browse all the source name belong to a specific source by click its name. For example, users can browse the families, genera or species belonging to fungi by click the organism "Fungi".
4. Browse by pathway
Under construction.
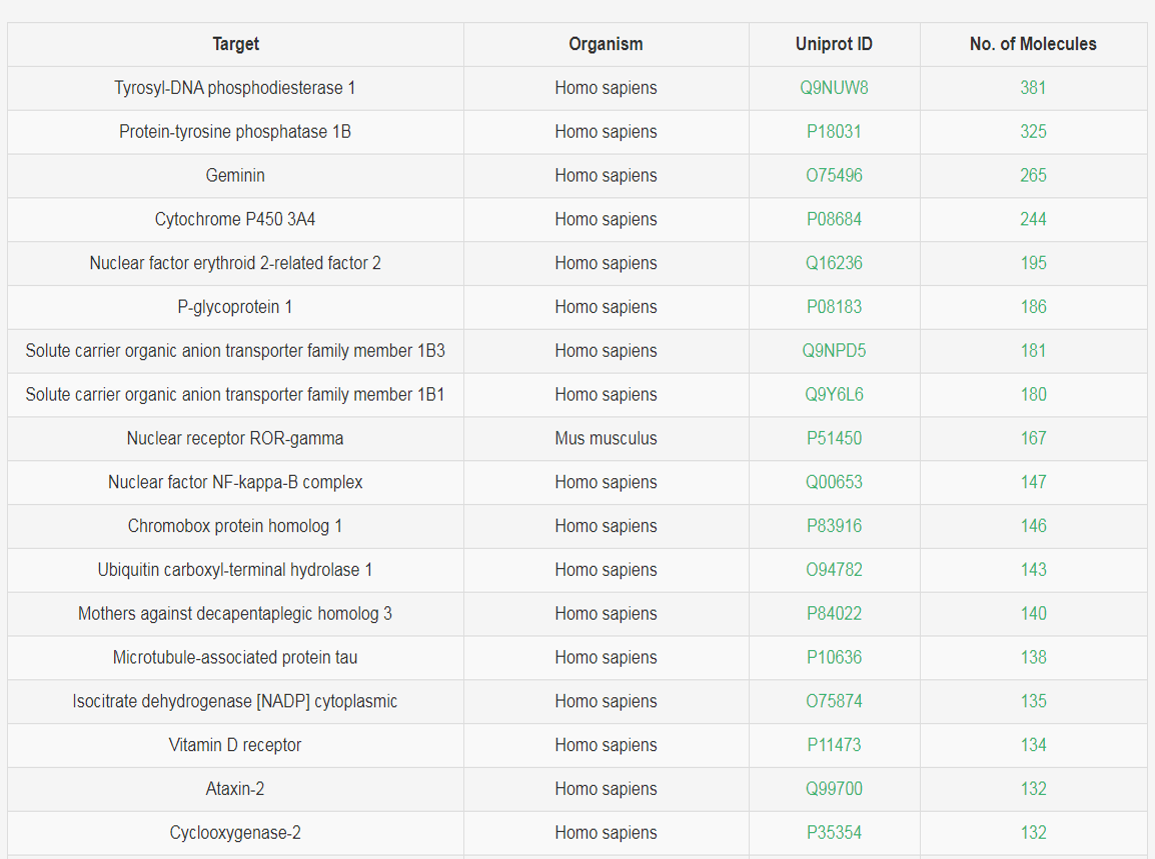
5. Browse by targets
The target name, organism and Uniprot ID are showed in the table, user can browse all compounds that act on the specific target by clicking the "No. of Molecules" column.
Search terokit
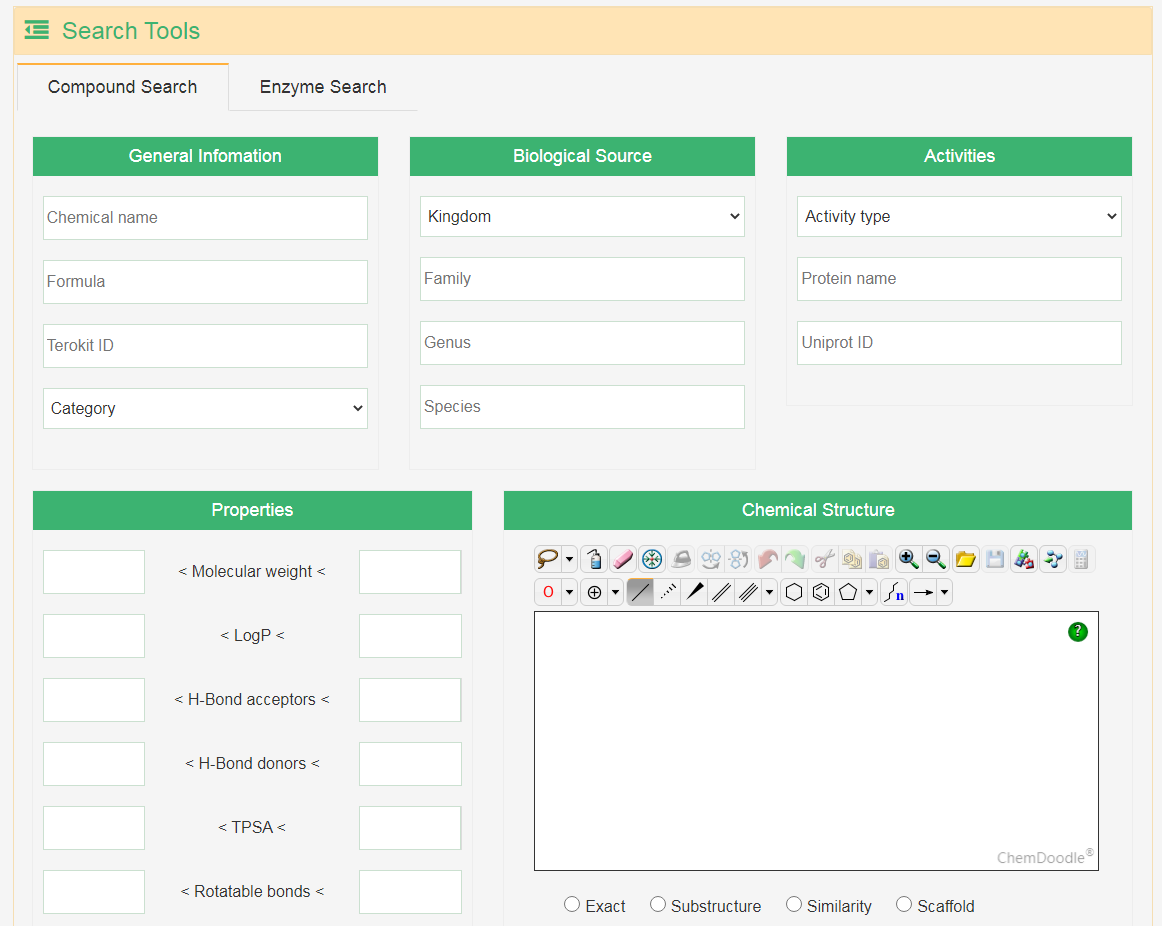
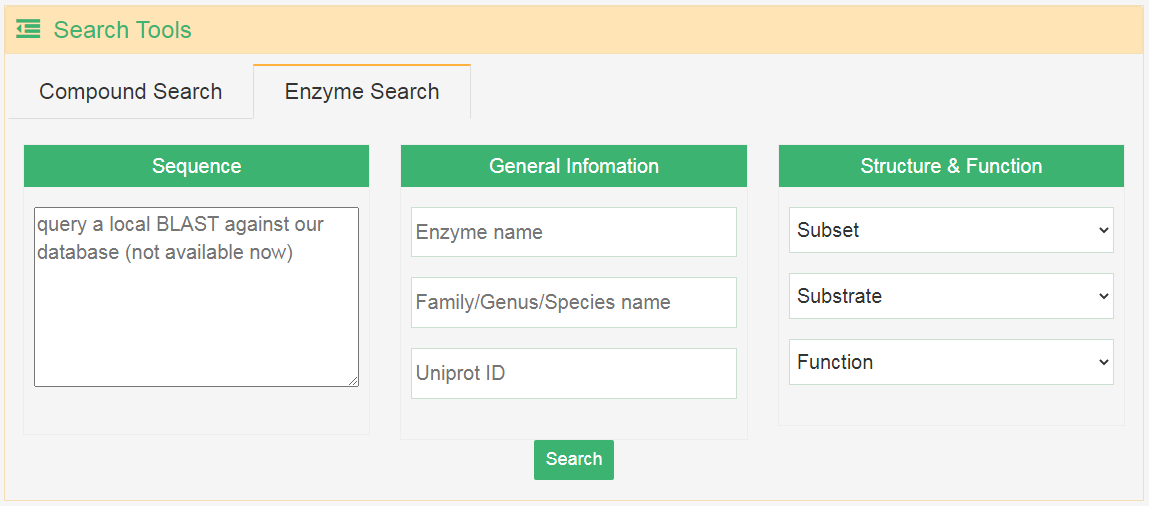
Users can input general information, biological source, activity information and properties to perform their search. Besides, users can also search for the compounds by drawing a structure, and the exact search, substructure search, similarity search and scaffold search are supported. The search for terpene synthases is also available.
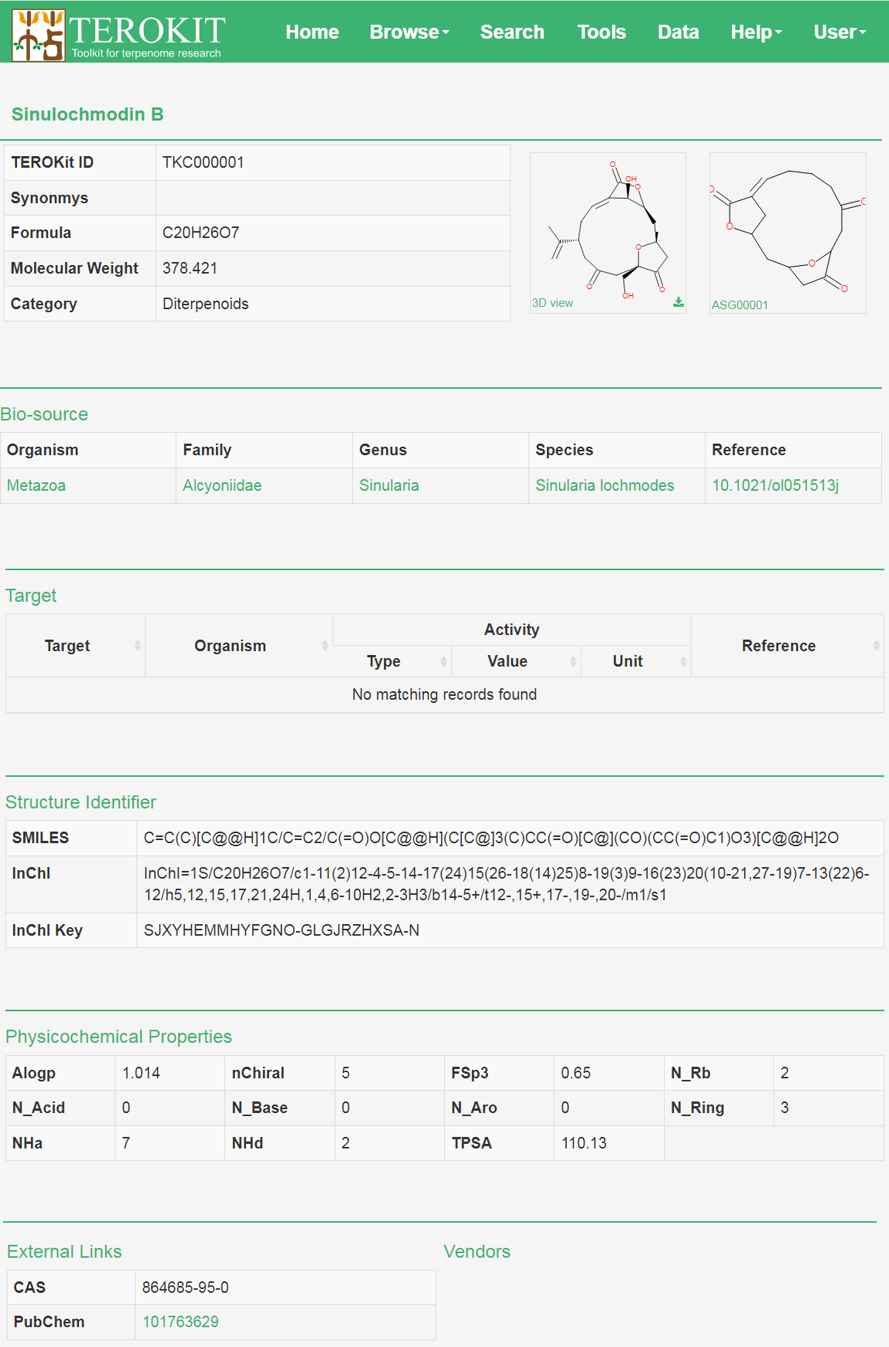
Compound page
All annotations about the compound is show in the compound page.
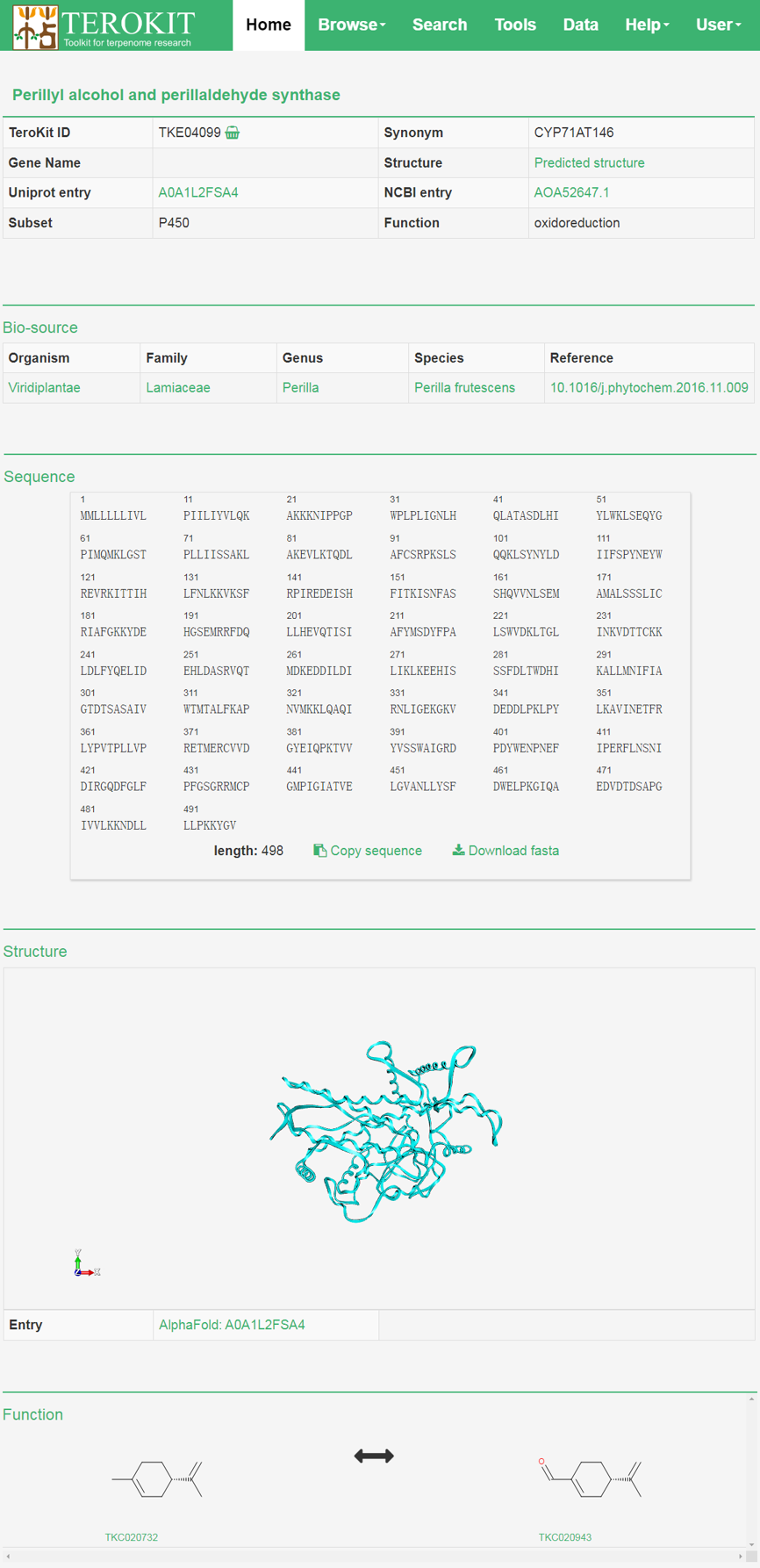
Enzyme page
All annotations about the enzyme is show in the enzyme page.
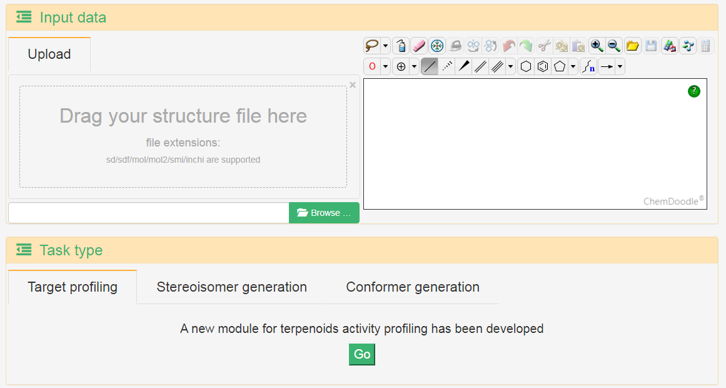
Terokit tools
Terokit provides some utilities in the tools page, including target prediction (visualization) and stereoisomers generation. Users can upload a structure file, paste the SMILES and InChI of a structure or draw a structure as the input.
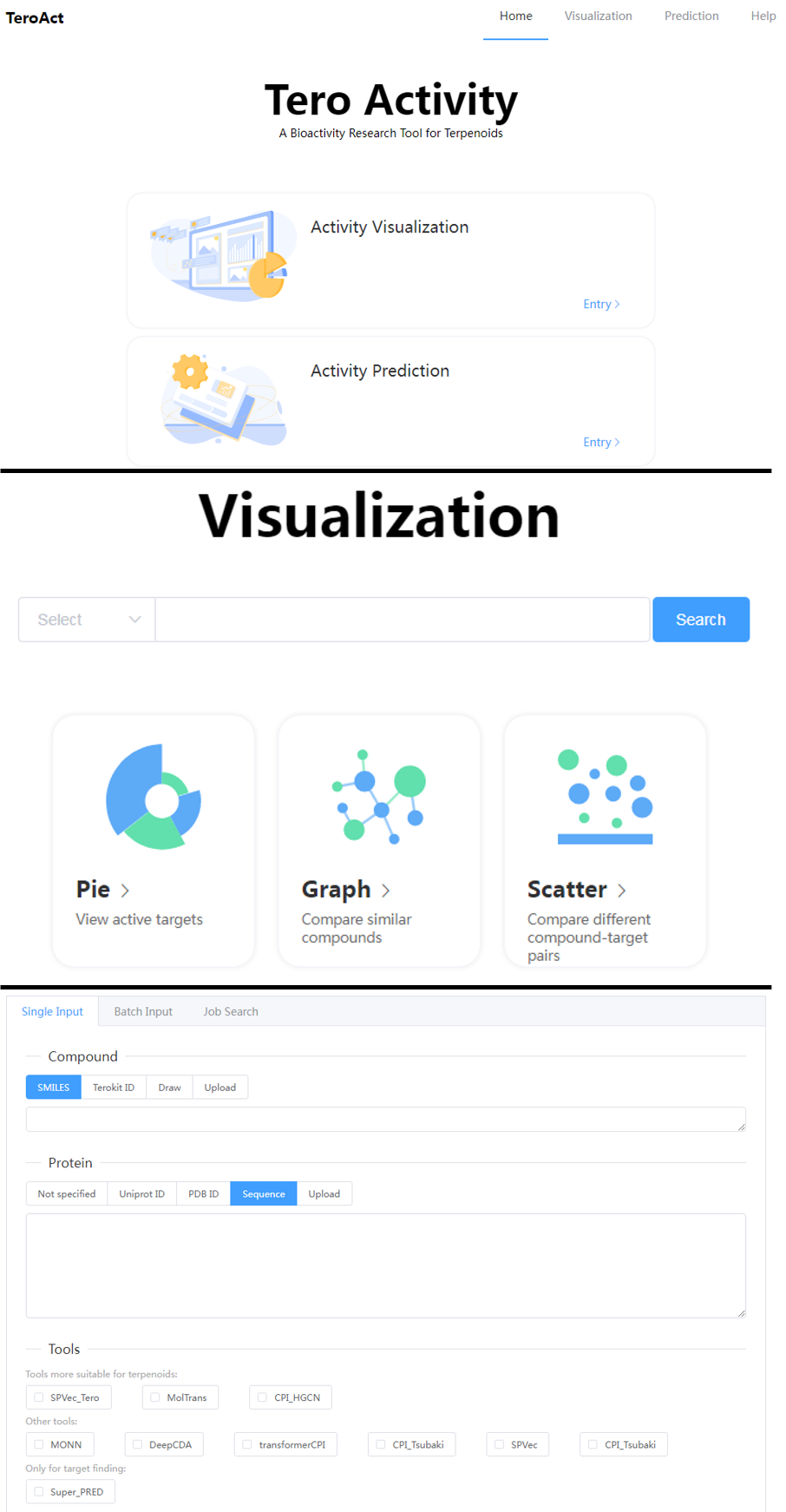
1. Target profiling
A independent module (TeroAct) is developed for the target visualization and prediction. All the compounds in TeroKit were matched with the data in ChEMBL and the activity information was collected. All biological activities can be visualized in kinds of charts. Several tools are integrated in the server for target prediction of terpenoids.
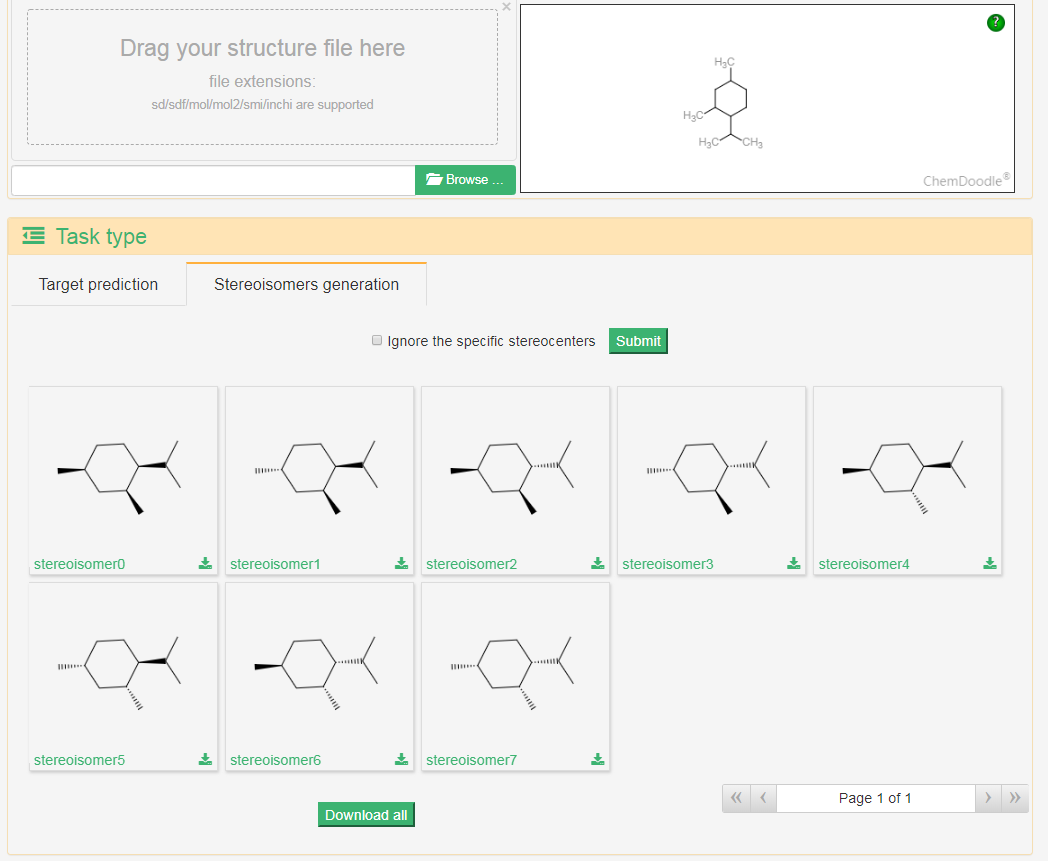
2. Stereoisomer generation
RDKit was used to generate the stereoisomers for a structure, users can also specify the stereochemistry of stereocenters in the molecule and TeroKit will not change them in the generation.
Data download and upload
Contents in TeroKit are listed in the download page, users can download all of them after registeration and login in. Data contribution is also welcome and appreciated, to upload page for more details about upload.
Citing TeroKit
TeroKit is free for academic use. Re-distribution of the data, in whole or in part, requires a license. For questions regarding TeroKit contents, licensing, or other support, please reach out to us by wurb3@mail.sysu.edu.cn
We ask that users who use TeroKit cite the papers:
Publication on the full TeroKit collection
Zeng, T.; Liu, Z.; Zhuang, J.; Jiang, Y.; He, W.; Diao, H.; Lv, N.; Jian, Y.; Liang, D.; Qiu, Y. ; Zhang, R.; Zhang, F.; Tang, X.; Wu, R. TeroKit: A Database-Driven Web Server for Terpenome Research. J. Chem. Inf. Model. 2020. DOI:10.1021/acs.jcim.0c00141
Publication on the TeroMOL database
Zeng, T.; Chen, Y.; Jian, Y.; Zhang, F.; Wu, R. Chemotaxonomic Investigation of Plant Terpenoids with an Established Database (TeroMOL). New Phytol. 2022 DOI:10.1111/nph.18133
Publication on the TeroENZ&TeroMAP database
Chen, N.; Zhang, R.; Zeng, T.; Zhang, X.; Wu, R. Developing TeroENZ and TeroMAP modules for the terpenome research platform TeroKit. Database 2023 DOI:10.1093/database/baad020